The Mg2+ binding sites of the 5S rRNA loop E motif as investigated by molecular dynamics simulations |
| |
| Authors: | Auffinger Pascal Bielecki Lukasz Westhof Eric |
| |
| Affiliation: | Institut de Biologie Moléculaire et Cellulaire du CNRS, Modélisations et Simulations des Acides Nucléiques, UPR 9002, 15 rue René Descartes, 67084 Cedex, Strasbourg, France. p.auffinger@ibmc.u-strasbg.fr |
| |
| Abstract: | 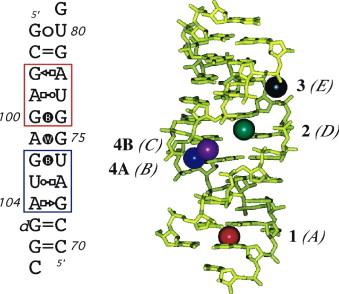
Molecular dynamics simulations have been used to investigate the binding of Mg(2+) ions to the deep groove of the eubacterial 5S rRNA loop E. The simulations suggest that long-lived and specific water-mediated interactions established between the hydrated ions and the RNA atoms lining up the binding sites contribute to the stabilization of this motif. The Mg(2+) binding specificity is modulated by two factors: (i) a required electrostatic complementarity and (ii) a structural correspondence between the hydrated ion and its binding pocket that can be estimated by its degree of dehydration and the resulting number and lifetime of the intervening water-mediated contacts. Two distinct binding modes for pentahydrated Mg(2+) ions that result in a significant freezing of the tumbling motions of the ions are described, and mechanistic details related to the stabilization of nucleic acids by divalent ions are provided. |
| |
| Keywords: | |
| 本文献已被 ScienceDirect PubMed 等数据库收录! |
|

