Evaluation of Digital Image Recognition Methods for Mass Spectrometry Imaging Data Analysis |
| |
| Authors: | Måns Ekelöf Kenneth P Garrard Rika Judd Elias P Rosen De-Yu Xie Angela D M Kashuba David C Muddiman |
| |
| Institution: | 1.FTMS Laboratory for Human Health Research, Department of Chemistry,North Carolina State University,Raleigh,USA;2.Department of Plant and Microbial Biology,North Carolina State University,Raleigh,USA;3.Division of Pharmacotherapy and Experimental Therapeutics,University of North Carolina at Chapel Hill,Chapel Hill,USA;4.Molecular Education, Technology, and Research Innovation Center (METRIC),North Carolina State University,Raleigh,USA |
| |
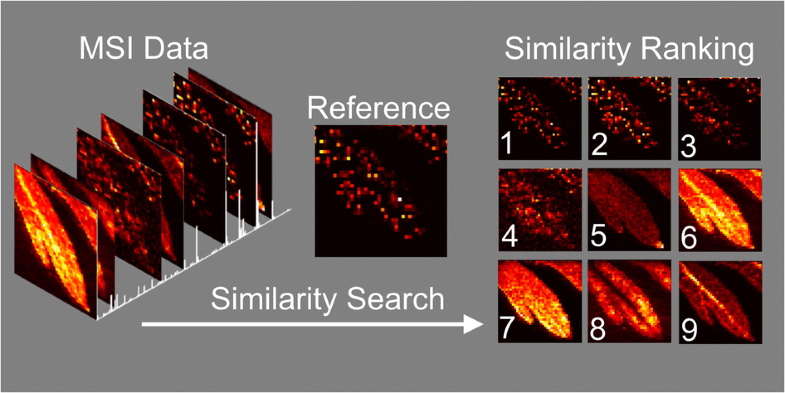
| Abstract: | Analyzing mass spectrometry imaging data can be laborious and time consuming, and as the size and complexity of datasets grow, so does the need for robust automated processing methods. We here present a method for comprehensive, semi-targeted discovery of molecular distributions of interest from mass spectrometry imaging data, using widely available image similarity scoring algorithms to rank images by spatial correlation. A fast and powerful batch search method using a MATLAB implementation of structural similarity (SSIM) index scoring with a pre-selected reference distribution is demonstrated for two sample imaging datasets, a plant metabolite study using Artemisia annua leaf, and a drug distribution study using maraviroc-dosed macaque tissue. |
| |
| Keywords: | |
| 本文献已被 SpringerLink 等数据库收录! |
|