|
|||||
|
|
Fluorometric determination of microRNA based on strand displacement amplification and rolling circle amplification |
|
| Authors: | Yunlei?Zhou Bingchen?Li Minghui?Wang |
| Institution: | 1.College of Chemistry and Material Science,Shandong Agricultural University,Taian,People’s Republic of China;2.College of Resources and Environment, Key Laboratory of Agricultural Environment in Universities of Shandong,Shandong Agricultural University,Taian,People’s Republic of China |
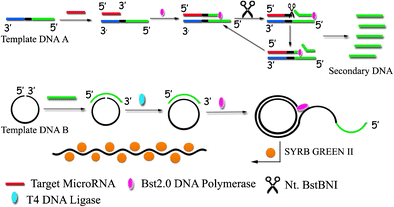
| Abstract: | The authors describe a fluorometric assay for microRNA. It is based on two-step amplification involving (a) strand displacement replication and (b) rolling circle amplification. The strand displacement amplification system is making use of template DNA (containing a sequence that is complementary to microRNA-21) and nicking enzyme sites. After hybridization, the microRNA strand becomes extended by DNA polymerase chain reaction and then cleaved by the nicking enzyme. The DNA thus produced acts as a primer in rolling circle amplification. Then, the DNA probe SYBR Green II is added to bind to ssDNA to generate a fluorescent signal which increases with increasing concentration of microRNA. The method has a wide detection range that covers the10 f. to 0.1 nM microRNA concentration range and has a detection limit as low as 1.0 fM. The method was successfully applied to the determination of microRNA-21 in the serum of healthy and breast cancer patients. Graphical abstract Schematic of a fluorometric microRNA assay based on two-step amplification involving strand displacement replication and rolling circle amplification. DNA probe SYBR Green II is then bound to ssDNA to generate a fluorescent signal which increases with increasing concentration of microRNA. |
| Keywords: | |
| 本文献已被 SpringerLink 等数据库收录! | |